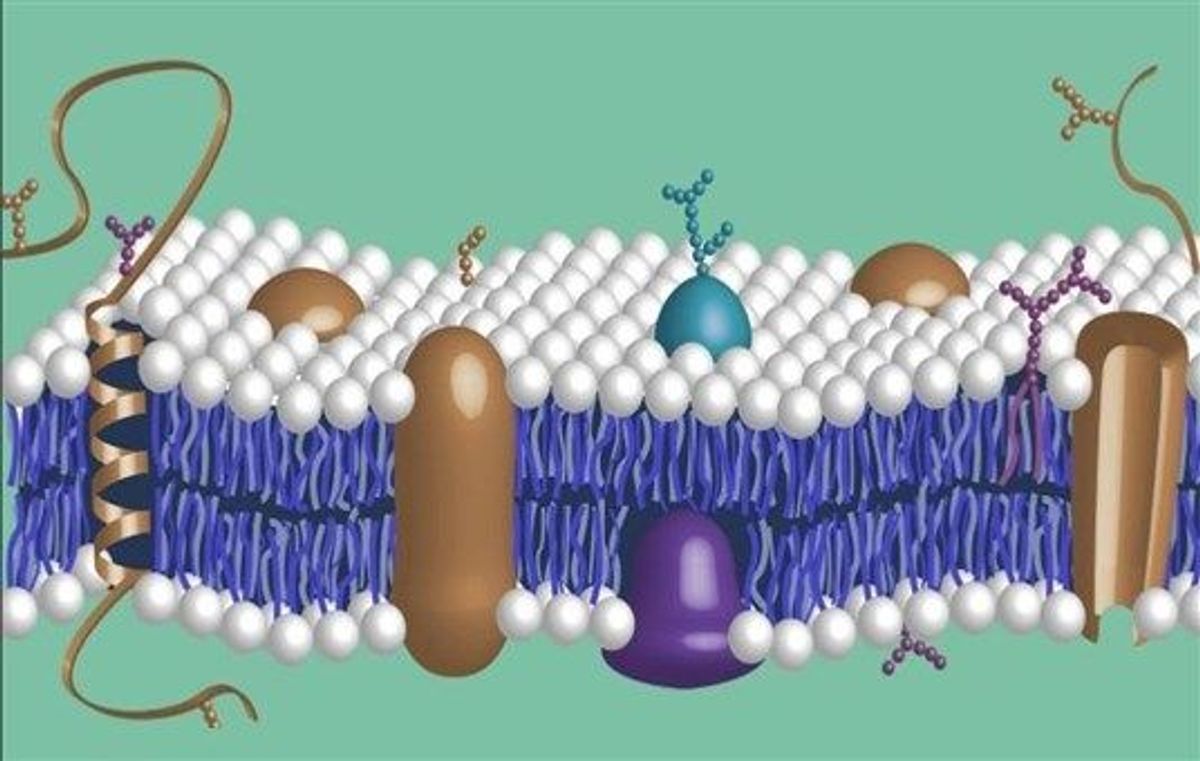
Spatial omics refers to a collection of technologies that allow researchers to map biological information to specific locations within a tissue sample. By combining genomic or proteomic profiling methods with spatial information, these techniques enable mapping molecular characteristics onto precise anatomy. This merging of spatial and molecular data provides insights into biology that standard bulk assays cannot reveal. Spatial omics overcomes the limitations of traditional methods by preserving molecular and cellular context and relationships that get lost through tissue homogenization.
Early Spatial Omics Technologies
Some of the earliest spatial omics methods involved in situ hybridization techniques, which detected specific RNA targets within intact tissues using fluorescently labeled probes. While useful for visualizing expression of a small number of genes, these methods lacked the throughput for comprehensive profiling. Another early approach was laser capture microdissection, which allowed isolating individual cells or groups of cells from histologically defined areas for subsequent molecular analysis. However, this disrupted spatial relationships and required prior knowledge of areas of interest.
Emergence of Spatial Transcriptomics
A major breakthrough was the development of Spatial Omics approaches that could profile gene expression across intact tissues with single-cell resolution. Spatial transcriptomics uses arrays of oligonucleotide probes linked to surfaces in ordered patterns. Tissue sections are placed on the arrays, allowing in situ hybridization to capture spatial information about thousands of RNA transcripts simultaneously. After imaging, the molecular data can be mapped back to specific locations on the slide. This non-destructive method preserves tissue architecture while providing spatially resolved transcriptomic profiles.
Advances in Spatial Proteomics
While transcriptomics gives insights into cellular states and functions, direct analysis of proteins is also invaluable due to post-transcriptional regulation. Spatial proteomics methods have accelerated incorporating protease-catalyzed protein tagging followed by sequencing or mass spectrometry. In one approach, tissue sections are treated with protease enzymes programmed to recognize and attach DNA-encoded tags to proteins at defined locations. After protein extraction, high-throughput sequencing identifies the spatially indexed protein signatures. Another strategy utilizes mass spectrometry imaging to directly map proteins within intact tissue sections based on their mass spectra. These emerging spatial proteomics technologies parallel the ability of spatial transcriptomics to profile molecular characteristics within anatomical context at high resolution.
Applications in Neuroscience and Immunology
Spatial Omics has provided novel insights across many areas of biology. In neuroscience, these methods have revealed molecular signatures associated with distinct brain cell types and helped characterize cellular diversity, connections, and disease states. Spatial transcriptomics of post-mortem brain tissue has uncovered gene expression patterns linked to psychiatric conditions like schizophrenia. Immunological studies have leveraged spatial omics to map immune cell infiltrates and molecular changes during infection, cancer, and autoimmunity within lymphoid organs and tumors. For example, profiling tumor-infiltrating immune cells has uncovered correlations between spatial distributions and clinical outcomes.
Engineering Advances Driving the Field Forward
Rapid engineering developments continue enhancing spatial omics technologies. Improved probe and antibody designs increase molecular coverage. Advances in tissue clearing and multiplexed labelling allow assaying additional classes of biomolecules within the same tissues. Combining spatial omics with in situ sequencing expands profiling to RNA variants and gene fusions. Automated image analysis facilitates extracting quantitative readouts from vast volumes of spatially registered data. Integrating genomic and epigenomic information provides deeper molecular context. Spatial omics is also being extended to model organisms via protocols preserving tissue architecture during multiplexed in situ analysis. These innovations make spatial omics an increasingly powerful approach for interrogating biology with unprecedented spatial and molecular resolution.
*Note:
1. Source: Coherent Market Insights, Public sources, Desk research
2. We have leveraged AI tools to mine information and compile it